
Protein Language Models (PLMs) have emerged as potent tools for predicting and designing protein structure and performance. On the International Conference on Machine Learning 2023 (ICML), MILA and Intel Labs released ProtST, a pioneering multi-modal language model for protein design based on text prompts. Since then, ProtST has been well-received within the research community, accumulating greater than 40 citations in lower than a yr, showing the scientific strength of the work.
Considered one of PLM’s hottest tasks is predicting the subcellular location of an amino acid sequence. On this task, users feed an amino acid sequence into the model, and the model outputs a label indicating the subcellular location of this sequence. Out of the box, zero-shot ProtST-ESM-1b outperforms state-of-the-art few-shot classifiers.
To make ProtST more accessible, Intel and MILA have re-architected and shared the model on the Hugging Face Hub. You possibly can download the models and datasets here.
This post will show you run ProtST inference efficiently and fine-tune it with Intel Gaudi 2 accelerators and the Optimum for Intel Gaudi open-source library. Intel Gaudi 2 is the second-generation AI accelerator that Intel designed. Take a look at our previous blog post for an in-depth introduction and a guide to accessing it through the Intel Developer Cloud. Because of the Optimum for Intel Gaudi library, you may port your transformers-based scripts to Gaudi 2 with minimal code changes.
Inference with ProtST
Common subcellular locations include the nucleus, cell membrane, cytoplasm, mitochondria, and others as described in this dataset in greater detail.
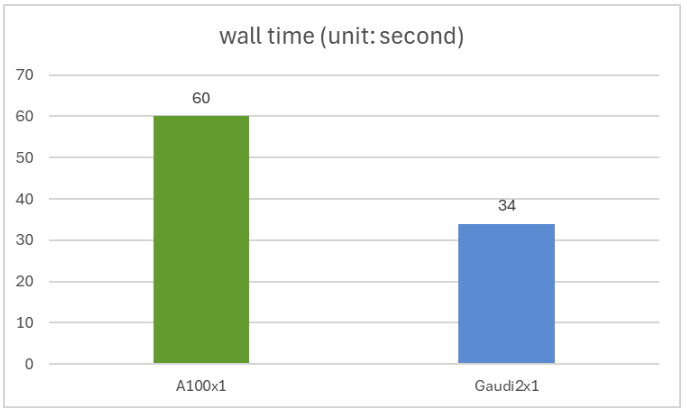
We compare ProtST’s inference performance on NVIDIA A100 80GB PCIe and Gaudi 2 accelerator using the test split of the ProtST-SubcellularLocalization dataset. This test set incorporates 2772 amino acid sequences, with variable sequence lengths starting from 79 to 1999.
You possibly can reproduce our experiment using this script, where we run the model in full bfloat16 precision with batch size 1. We get an analogous accuracy of 0.44 on the Nvidia A100 and Intel Gaudi 2, with Gaudi2 delivering 1.76x faster inferencing speed than the A100. The wall time for a single A100 and a single Gaudi 2 is shown within the figure below.
Superb-tuning ProtST
Superb-tuning the ProtST model on downstream tasks is a straightforward and established solution to improve modeling accuracy. On this experiment, we specialize the model for binary location, an easier version of subcellular localization, with binary labels indicating whether a protein is membrane-bound or soluble.
You possibly can reproduce our experiment using this script. Here, we fine-tune the ProtST-ESM1b-for-sequential-classification model in bfloat16 precision on the ProtST-BinaryLocalization dataset. The table below shows model accuracy on the test split with different training hardware setups, and so they closely match the outcomes published within the paper (around 92.5% accuracy).

The figure below shows fine-tuning time. A single Gaudi 2 is 2.92x faster than a single A100. The figure also shows how distributed training scales near-linearly with 4 or 8 Gaudi 2 accelerators.

Conclusion
On this blog post, we now have demonstrated the benefit of deploying ProtST inference and fine-tuning on Gaudi 2 based on Optimum for Intel Gaudi Accelerators. As well as, our results show competitive performance against A100, with a 1.76x speedup for inference and a 2.92x speedup for fine-tuning.
The next resources will enable you to start together with your models on the Intel Gaudi 2 accelerator:
Thanks for reading! We stay up for seeing your innovations built on top of ProtST with Intel Gaudi 2 accelerator capabilities.
